
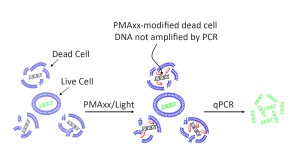
PMAxx™dyeisaDNAmodifierusedforviabilityPCR,inventedbyscientistsatBiotium.PMAxx™isanewandimprovedversionofourpopularviabilitydyePMA(propidiummonoazide).LikePMA,PMAxx™isaphoto-reactivedyethatbindstodsDNAwithhighaffinity.UponphotolysiswithvisIBLelight,PMAxx™dyebecomescovalentlyattachedtodsDNA.ThePMAxx™-modifieddsDNAcannotbeamplifiedbyPCR.PMAxx™dyeisdesignedtobecellmembrane-impermeable.Thus,inapopulationofliveanddeadcells,onlydeadcellsaresusceptibletoDNAmodificationduetocompromisedcellmembranes.ThisuniquefeaturemakesPMAxx™highlyusefulinselectivedetectionoflivebacteriabyqPCR.
PMAxx™wasdesignedbyBiotiumscientiststobeasuperioralternativetoPMA.WhilePMAisgenerallyeffectiveatdifferentiatingbetweenliveanddeadbacteriabyqPCR,itdoesnotcompletelyeliminatePCRproductsfromdeadcell DNA.Thiscouldpotentiallygivefalsepositiveresults.Biotium’snewdyePMAxx™ismuchmoreeffectiveateliminatingPCRamplificationofdeadcellDNA,andthereforeprovidesthebest discriminationbetweenliveanddeadbacteria.
SinceBiotiumfirstdevelopedPMAdye,therehavebeennumerouspublicationsontheuseofthedyeinpathogenicbacterialdetectionrelatedtofoodandwatersafety,medicaldiagnosisandbiodefense(downloadthePMAReferenceList).PMAxx™shouldbecompatiblewithallexistingPMAprotocols,butwithsuperioractivity.OfcoursewealsostillselltheclassicPMAdye,inwater(40019)orlyophilizedformat(40013).
ForviabilityPCRofGram-negativebacteriawehighlyrecommendusingourPMAEnhancer(31038)inconjunctionwithPMAxx™.PMAEnhancerforGramNegativeBacteriawasdesignedtoimprovePMA-mediateddiscriminationbetweenliveanddeadGram-negativebacteria.WhenasequencefromaGram-negativebacteriaisamplifiedbyPCR,samplespre-treatedwithdyeandEnhancershowadecreaseinthesignalfromdeadcells,withnochangeinthesignalfromlivecells.PMAEnhancerisparticularlyusefulforsamplesinwhichbacteriawerekilledusingmildmethodssuchaslowheat.
Biotiumalsoprovidesstrain-specificPMAReal-TimePCRBacterialViabilitykitswithvalidatedprimersfor7selectedpathogens:M.tuberculosis,S.aureus,MRSA,Salmonella,E.coli,E.coliO157:H7andListeria.Thesekitsprovideeverythingthatyouneedfortheselectivedetectionofyourfavoritespeciesoflivebacteriabyreal-timePCR.Don’tseeyourfavoritestrain?Letusknowattechsupport@biotium.com.
Real-TimePCRBacterialViabilityKits
ForphotoactivationofPMAxx™dye(orPMAorEMA),werecommendtheuseofBiotium’sPMA-Lite™LEDPhotolysisDevice(E90002),whichisdesignedtoconductphotolysisundercontrolledconditions.
- Abs=466nm(beforephotolysis)
- Abs/Em=~510/~610nm(followingphotolysisandcovalentattachmenttoDNA/RNA)
- Darkredsolution
- Storeat-20°Candprotectfromlightatalltimes
References
PMAxxReferences
Garcia-Fontana,C.,etal.(2016)ANewPhysiologicalRolefortheDNAMoleculeasaProtectoragainstDryingStressinDesiccation-TolerantMicroorganisms.Front.Microbiol.Dec22;7:2066.
Randazzo,W.,etal.(2016)EvaluationofviabilityPCRperformanceforassessingnorovirusinfectivityinfresh-cutvegetablesandirrigationwater.Int.J.FoodMicrobiol.Jul16;229:1-6.
Randazzo,W.,etal.(2017).Improvingefficiencyofviability-qPCRforselectivedetectionofinfectiousHAVinfoodandwatersamples.JApplMicrobiol.AcceptedAuthorManuscript.doi:10.1111/jam.13519
PMAReferences
Nocker,A.,Cheung,C.Y.,andKamper,A.K.(2006).Comparisonofpropidiummonoazidewithethidiummonoazidefordifferentiationoflivevs.deadbacteriabyselectiveremovalofDNAfromdeadcells.J.MicrobioMeth.67(2),310-320.
DownloadthefullPMAReferenceList.



